Heterogeneities in Axonal Structure and Transporter Distribution Lower Dopamine Reuptake Efficiency
Abstract
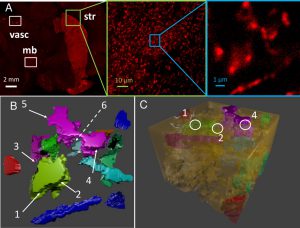
Efficient clearance of dopamine (DA) from the synapse is key to regulating dopaminergic signaling. This role is fulfilled by DA transporters (DATs). Recent advances in the structural characterization of DAT from Drosophila (dDAT) and in high-resolution imaging of DA neurons and the distribution of DATs in living cells now permit us to gain a mechanistic understanding of DA reuptake events in silico. Using electron microscopy images and immunofluorescence of transgenic knock-in mouse brains that express hemagglutinin-tagged DAT in DA neurons, we reconstructed a realistic environment for MCell simulations of DA reuptake, wherein the identity, population and kinetics of homology-modeled human DAT (hDAT) sub-states were derived from molecular simulations. The complex morphology of axon terminals near active zones was observed to give rise to large variations in DA reuptake efficiency, and thereby in extracellular DA density. Comparison of the effect of different firing patterns showed that phasic firing would increase the probability of reaching local DA levels sufficiently high to activate low-affinity DA receptors, mainly due to high DA levels transiently attained during the burst phase. The experimentally observed non-uniform surface distribution of DATs emerged as a major modulator of DA signaling: reuptake was slower, and the peaks/width of transient DA levels were sharper/wider under non-uniform distribution of DATs, compared to uniform. Overall, the study highlights the importance of accurate descriptions of extra-synaptic morphology, DAT distribution and conformational kinetics for quantitative evaluation of dopaminergic transmission and for providing deeper understanding of the mechanisms that regulate DA transmission.
